
MASS SPECTROMETRY
Mass Spec Services Overview
BGI Americas now offers proteomic and biologics characterization services to accelerate your life science research program. Our new portfolio of end-to-end LC-MS services leverages BGI’s strengths in managing large scale computing infrastructure and pioneering innovative bioinformatics technologies.
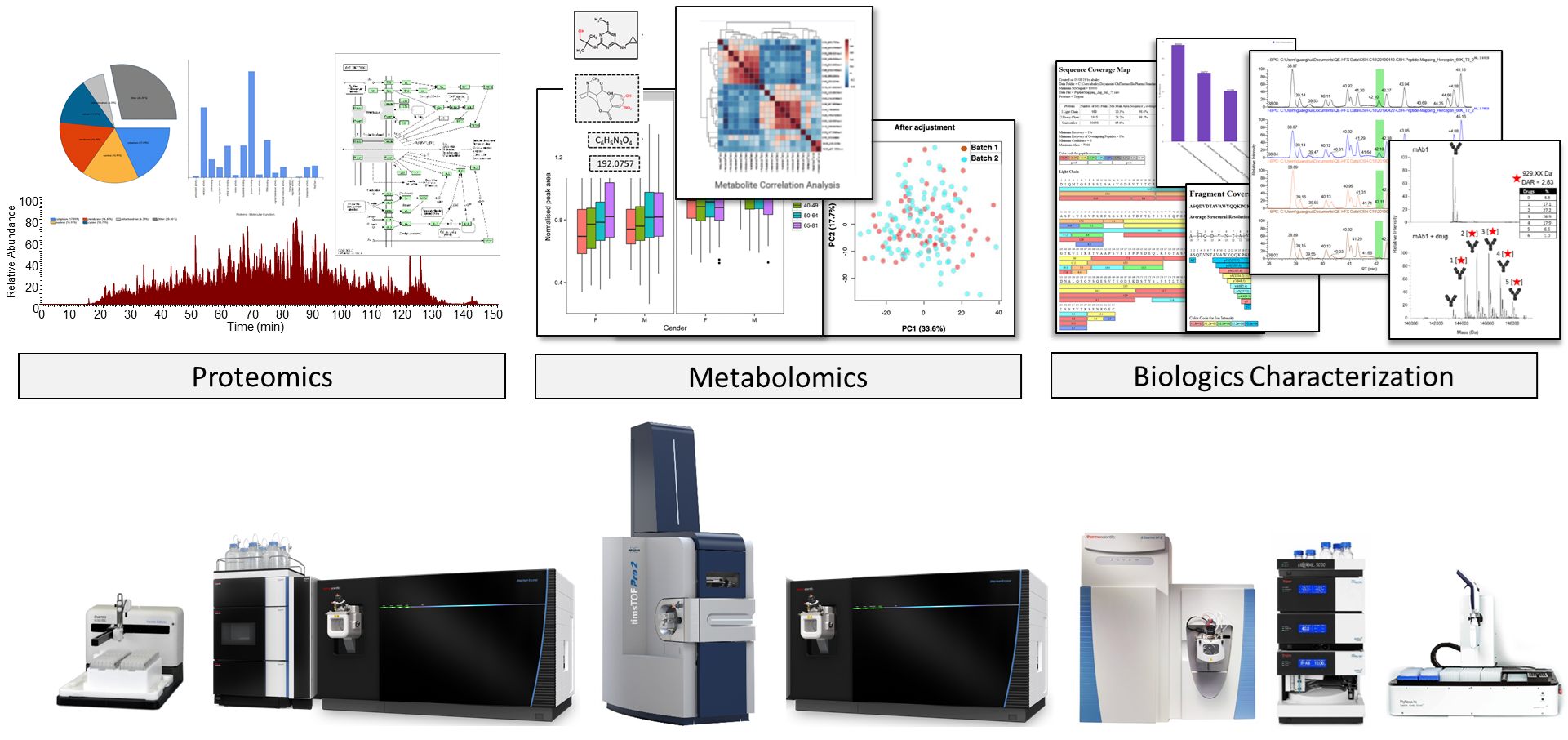
Our mass spectrometry service laboratory in San Jose, CA is comprised of scientists with extensive experience in liquid chromatography and mass spectrometry-based analytical methods. This state-of-the-art facility is designed to support a broad range of protein research applications.
We can help simplify your research challenge. BGI can provide workflow customization and consultation services to address unique project requirements. Turnaround time is 1 or 2 weeks for certain premium services.
Proteomic Services
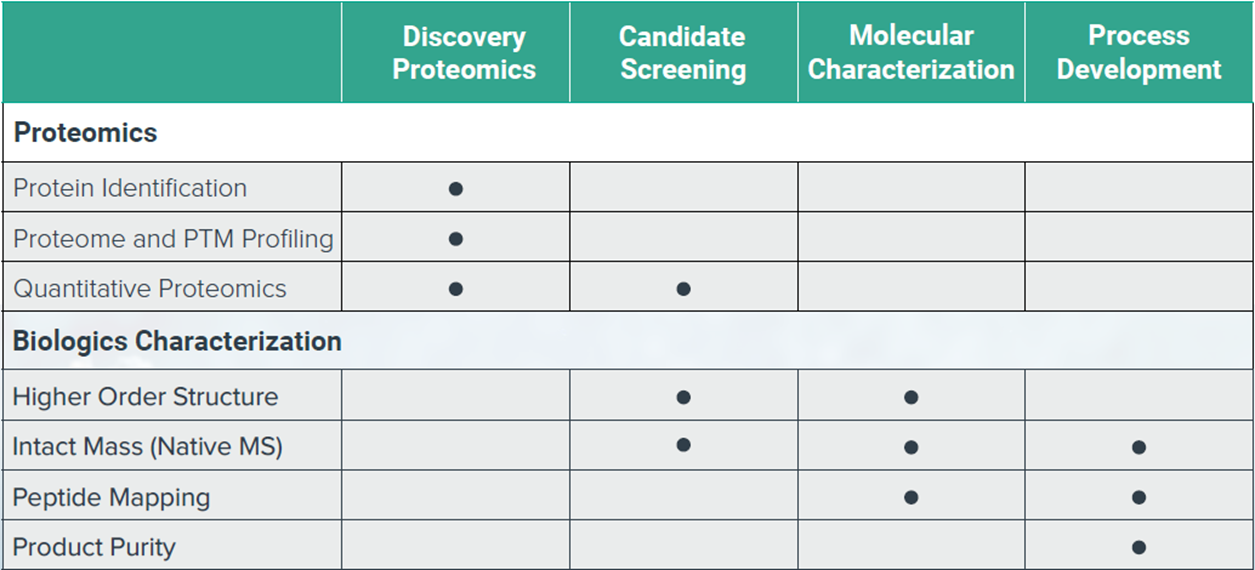
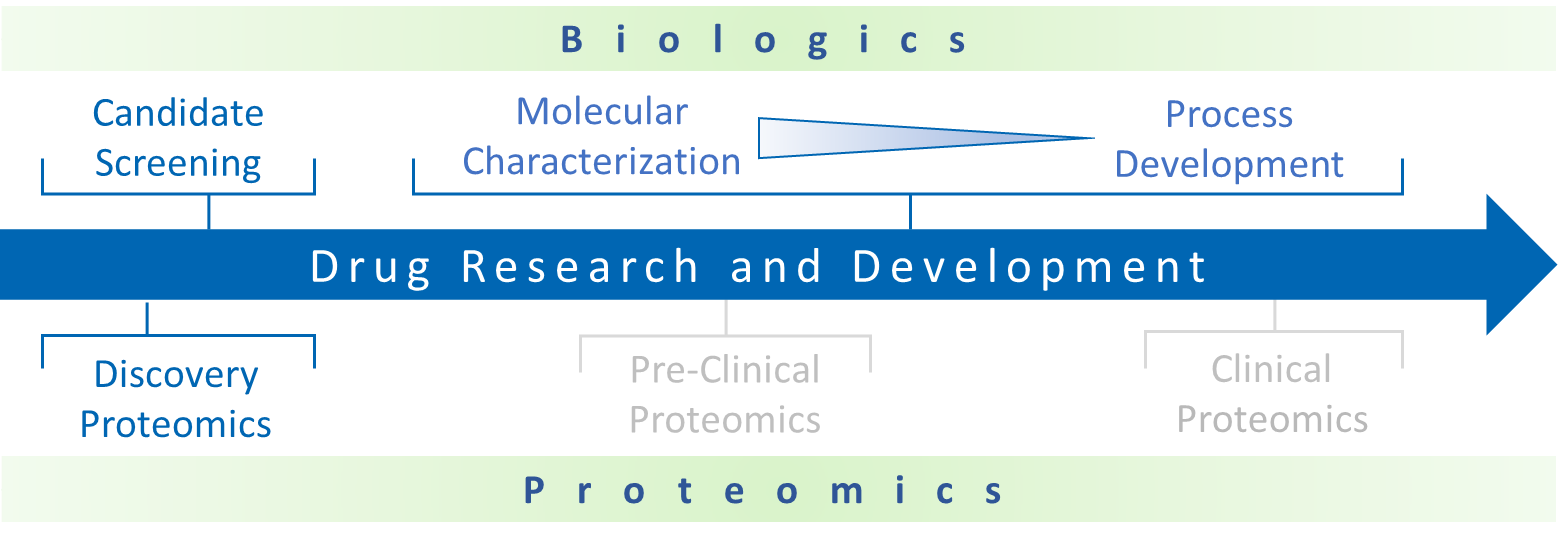
BGI provides global and targeted solutions for protein quantitation1,2. We can flexibly adapt our Label-Free DIA and Isobaric Label TMT services to best fit your experimental strategies3,4,5. For screening applications we provide a highly specific targeted peptide quantitation using Parallel Reaction Monitoring (PRM) scanning using high resolution Orbitrap MS detection6.
We offer services for large scale proteomic profiling and can provide sample pre-fractionation using our off-line UHPLC-UV platform. Our laboratory can also perform Protein Identification services using solution samples or gel-isolated samples.
All of our proteomic services can be customized to meet your drug discovery project needs.
Biologics Characterization Services
BGI scientists in San Jose, California, provide analytical services and downstream support for biopharmaceutical and biotechnology industry customers. Our goal is to provide you with detailed knowledge about the molecular composition of your biologic sample using innovative analytical methods optimized for dynamic range and sensitivity.
Our state-of-the-art LC-MS platform can support diverse biologic characterization projects, including highly complex samples such as bi-/multi-specific antibodies, ADCs, and Fc-fusion protein drugs. Additionally, our scientific team at the San Jose Mass Spectrometry Center have pioneered native LC-MS technology which can preserve non-covalent binding and is compatible with medium-high sample throughput7,8,9.
We can provide broad support for drug discovery and other biologic-related proteomics services, including phosphoproteomics, CAR-T interactomics, and trans-omics projects involving proteomic and transcriptomic (DNBseq) analyses.
References
[1]. Xun Z, Shangbo X et al. Tissue-specific Proteogenomics Analysis of Plutella xylostella Larval Midgut Using a Multialgorithm Pipeline. Mol Cell Proteomics. 2016; 15(6): 1791-1807. doi: 10.1074/mcp.M115.050989.
[2]. Zhen C, Bo W et al. Quantitative proteomics reveals the temperature-dependent proteins encoded by a series of cluster genes in thermoanaerobacter tengcongensis. Mol Cell Proteomics. 2013; 12(8): 2266-2277. doi: 10.1074/mcp.M112.025817.
[3]. Wong, J. W. H., & Cagney, G. (2009). An Overview of Label-Free Quantitation Methods in Proteomics by Mass Spectrometry. Proteome Bioinformatics, 273–283.doi:10.1007/978-1-60761-444-9_18.
[4]. Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX, Villén J, MacCoss MJ. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun. 2018 Dec 3;9(1):5128. doi: 10.1038/s41467-018-07454-w.
[5]. Kelstrup CD, Bekker-Jensen DB, Arrey TN, Hogrebe A, Harder A, Olsen JV. Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J Proteome Res. 2018 Jan 5;17(1):727-738. doi: 10.1021/acs.jproteome.7b00602.
[6]. Bourmaud A, Gallien S, Domon B. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. Proteomics. 2016 Aug;16(15-16):2146-59. doi: 10.1002/pmic.201500543.
[7]. Schachner L, Han G, Dillon M, Zhou J, McCarty L, Ellerman D, Yin Y, Spiess C, Lill JR, Carter PJ, Sandoval W. Characterization of Chain Pairing Variants of Bispecific IgG Expressed in a Single Host Cell by High-Resolution Native and Denaturing Mass Spectrometry. Anal Chem. 2016 Dec 20;88(24):12122-12127. doi:10.1021/acs.analchem.6b02866.
[8]. Charge variant native mass spectrometry benefits mass precision and dynamic range of monoclonal antibody intact mass analysis. Bailey AO, Han G, Phung W, Gazis P, Sutton J, Josephs JL, Sandoval W. MAbs. 2018 Nov-Dec;10(8):1214-1225. doi: 10.1080/19420862.2018.1521131.
[9]. Ren C, Bailey AO, VanderPorten E, Oh A, Phung W, Mulvihill MM, Harris SF, Liu Y, Han G, Sandoval W. Quantitative Determination of Protein-Ligand Affinity by Size Exclusion Chromatography Directly Coupled to High-Resolution Native Mass Spectrometry. Anal Chem. 2019 Jan 2;91(1):903-911. doi:10.1021/acs.analchem.8b03829.